Concepción González-Bello
Universidade de Santiago de Compostela, Spain
Title: Disabling essential enzyme motion for catalysis: An efficient approach for shikimate kinase inhibition
Biography
Biography: Concepción González-Bello
Abstract
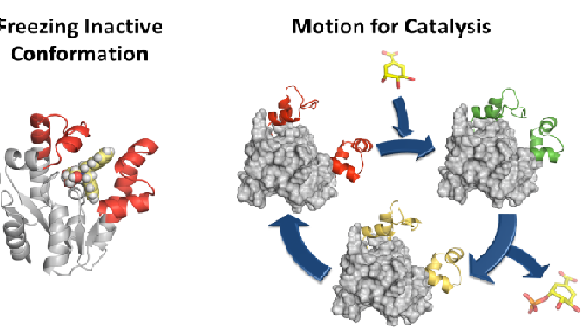
Although antibiotics are one of the most successful drugs in clinic that have saved millions of lives, many of them are nowadays ineffective in treating infections caused by resistant bacteria. It is therefore urgent to search for new antibacterial agents and approaches to face this huge challenge. Considering that most current antibiotics that are highly successful in human clinical use, targeted at only four main key processes and resistance to these antibiotics is widespread and well known, the search for unexplored bacterial functions appears to be a good option for the development of novel antimicrobial agents with a new mechanism of action. Our group is studying the possible development of new antibiotics by the selective and effective inhibition of an essential enzyme in bacteria that does not have any counterpart in human cells, shikimate kinase (SK, aroK gene). In particular, we are focused on SK from M. tuberculosis and H. pylori, two important pathogenic bacteria. Based on the essential enzyme motion for catalysis and product release studied by molecular dynamics simulation studies, potent reversible competitive inhibitors of the enzyme were developed. Compounds that stabilize the closed conformation for catalysis or the open conformation for product release were developed. An ester prodrug approach was used for achieving good in vitro activities against H. pylori. Our results also show that the less exploited motion-based design approach, not only is an alternative strategy for the development of competitive inhibitors, but could also be a way to achieve selectivity against a particular enzyme among its homologous ones. By using this approach, (1) the presence in the selected pocket of residues with markedly different properties would not be required, and (2) the effects of changes on residues to avoid the inhibition (resistance) should have a less pronounced effect.
Recent Publications
1.Blanco B, Prado V, Lence E, Otero J M, Garcia-Doval C, van Raaij M J, Llamas-Saiz A L, Lamb H, Hawkins A R, González-Bello C (2013) Mycobacterium tuberculosis shikimate kinase inhibitors: Design and simulation studies of the catalytic turnover. J. Am. Chem. Soc. 135: 12366-12376.
2.Prado V, Lence E, Maneiro M, Vázquez-Ucha J C, Beceiro A, Thompson P, Hawkins A R, González-Bello C (2016) Targeting the motion of shikimate kinase: Development of competitive inhibitors that stabilize an inactive open conformation of the enzyme. J. Med. Chem. 59: 5471-5487.
3.Prado V, Lence E, Thompson P, Hawkins A R, González-Bello C (2016) Freezing the dynamic gap for selectivity - motion-based design of inhibitors of the shikimate kinase enzyme. Chem. Eur. J. 22: 17988-18000.
4.Prado V, Lence E, Vallejo J A, Beceiro A, Thompson P, Hawkins A R, González-Bello C (2016) Study of the Phosphoryl-transfer mechanism of shikimate kinase by NMR spectroscopy. Chem. Eur. J. 22: 2758-2768.
5.González-Bello C (2016) Inhibition of shikimate kinase and type II dehydroquinase for antibiotic discovery: Structure-based design and simulation studies. Curr. Top Med. Chem. 16: 960-977.